After screening out the compounds showing potential activity against the target through the hit identification stage, it is necessary to carry out further characterization and optimization of these hit compounds. And this stage is termed as hit to lead. The goal of this stage is to identify the most promising hit series through limited structure-activity relationship (SAR) studies to enter the lead optimization and preclinical development stage. With a dedicated team of skilled scientists and technicians, Creative Biostructure provides MagHelix™ hit evaluation and lead identification (hit to lead) solutions to meet our clients’ needs and goals.
Whether your therapeutic goal focuses on the development of small molecules, peptides, or other biologics, Creative Biostructure can ensure that the performance of your initial hits is significantly improved to meet the standards required to enter the lead optimization stage. We can prioritize and classify the hit compounds identified through screening platforms and utilize advanced biophysical techniques to analyze the target-ligand binding modes. In addition, computer-aided drug design (CADD) tools can help you perform pharmacophore modeling, QSAR analysis, ADME/T prediction, etc. The molecules after the hit to lead stage are the most likely potential leads, which have enhanced potency, reduced off-target activity, and predicted physiochemical/metabolic properties of reasonable in vivo pharmacokinetics.
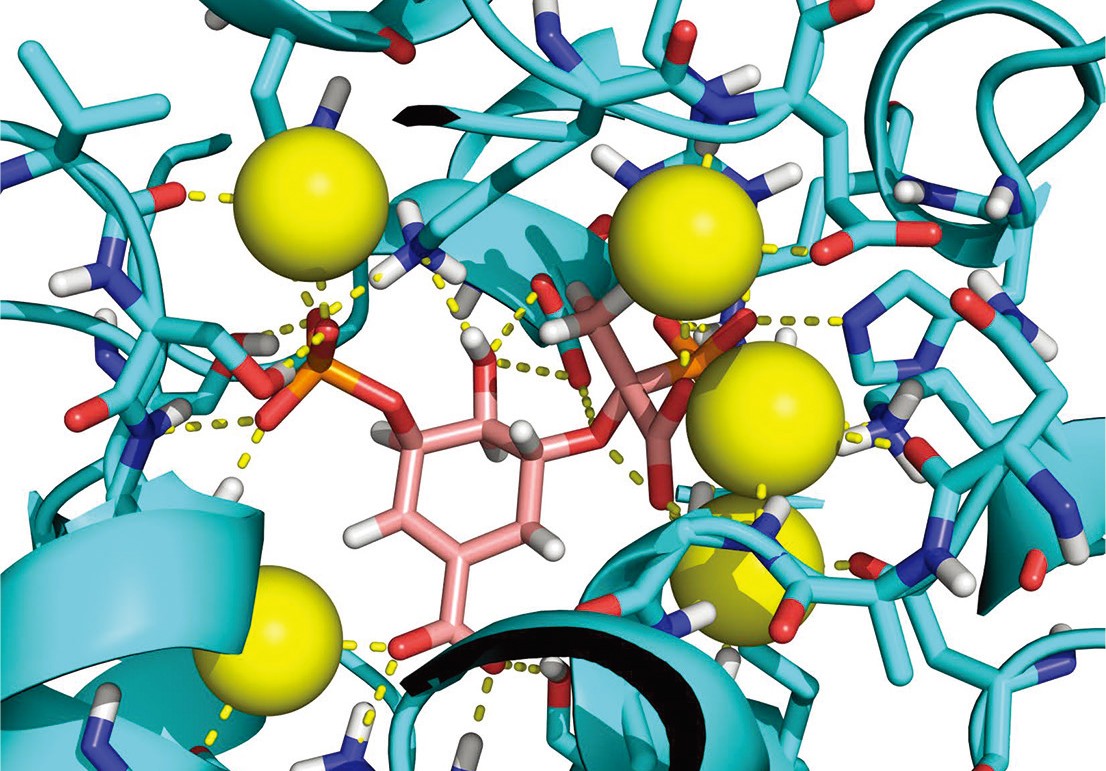
As drug discovery enters the hit to lead stage, more detailed information on hit compounds is needed to guide the choice of the best compounds for the next step. Creative Biostructure provides a variety of mainstream biophysical techniques for the identification and characterization of hit compounds. Information obtained can range from simply determining whether the ligand interacts with the target, to binding specificity and information about the binding mode. For example, biophysical assays detect the binding affinities between a hit and the target by Nuclear Magnetic Resonance (NMR), Isothermal Titration Calorimetry (ITC), Dynamic Light Scattering (DLS), and Surface Plasmon Resonance (SPR) techniques. We can also obtain structural information of target-ligand complexes and characterize the binding sites by assays such as X-ray crystallography, Cryo-electron microscopy (Cryo-EM), and NMR. These data are the basis for lead compound discovery and optimization.
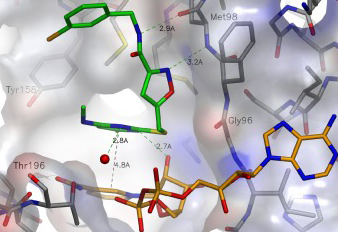
Fragment-based drug discovery (FBDD) has become a successful key technology for early drug discovery and development in the pharmaceutical industry. The low molecular weight compounds obtained through fragment screening can bind at one or more sites on the macromolecular target and serve as starting points for the development of leads. With rich experience and expertise in the field of drug design, Creative Biostructure can provide comprehensive fragment-to-lead solutions. We utilize fragment growth, fragment linking, and fragment merging strategies to optimize the preliminarily screened fragments to improve their biological activity and drug-like properties, making it a potential lead compound with higher affinity.
Not only can we offer fragment-to-lead solutions, but we can also support upstream fragment-based screening (FBS) and downstream lead optimization. Moreover, we welcome scientists from academia, biotech, and pharmaceutical industries to use our FBDD platform to implement projects related to drug discovery.
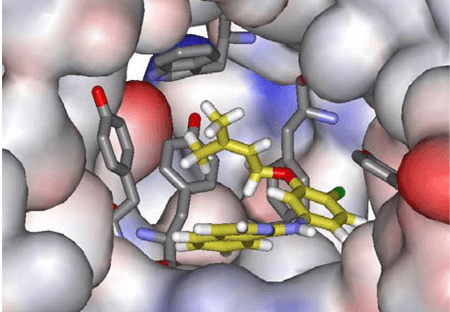
De novo drug design is a virtual hit to lead approach that automatically generates putative optimized compounds “from scratch” within the binding site of known 3D structures using predefined sets of substructures and rules governing their linking. De novo drug design can complement high-throughput screening (HTS) and virtual screening, and this approach can explore considerably larger chemical space. Creative Biostructure can use this computational technique to generate novel molecules or molecular scaffolds with predictive pharmacological properties fromscratch. Based on the availability of the 3D structure of the target protein, we can adopt two strategies,namely structure-based de novo drug design (SBDND) and ligand-based de novo drugdesign (LBDND). Our virtual hit to lead approach relying on the de novo design has been upgradedto utilize a pre-positioned fragment in the binding site as the original seed. Starting from experimentallyvalidated fragments with known binding modes may improve the reliability of their predictions.
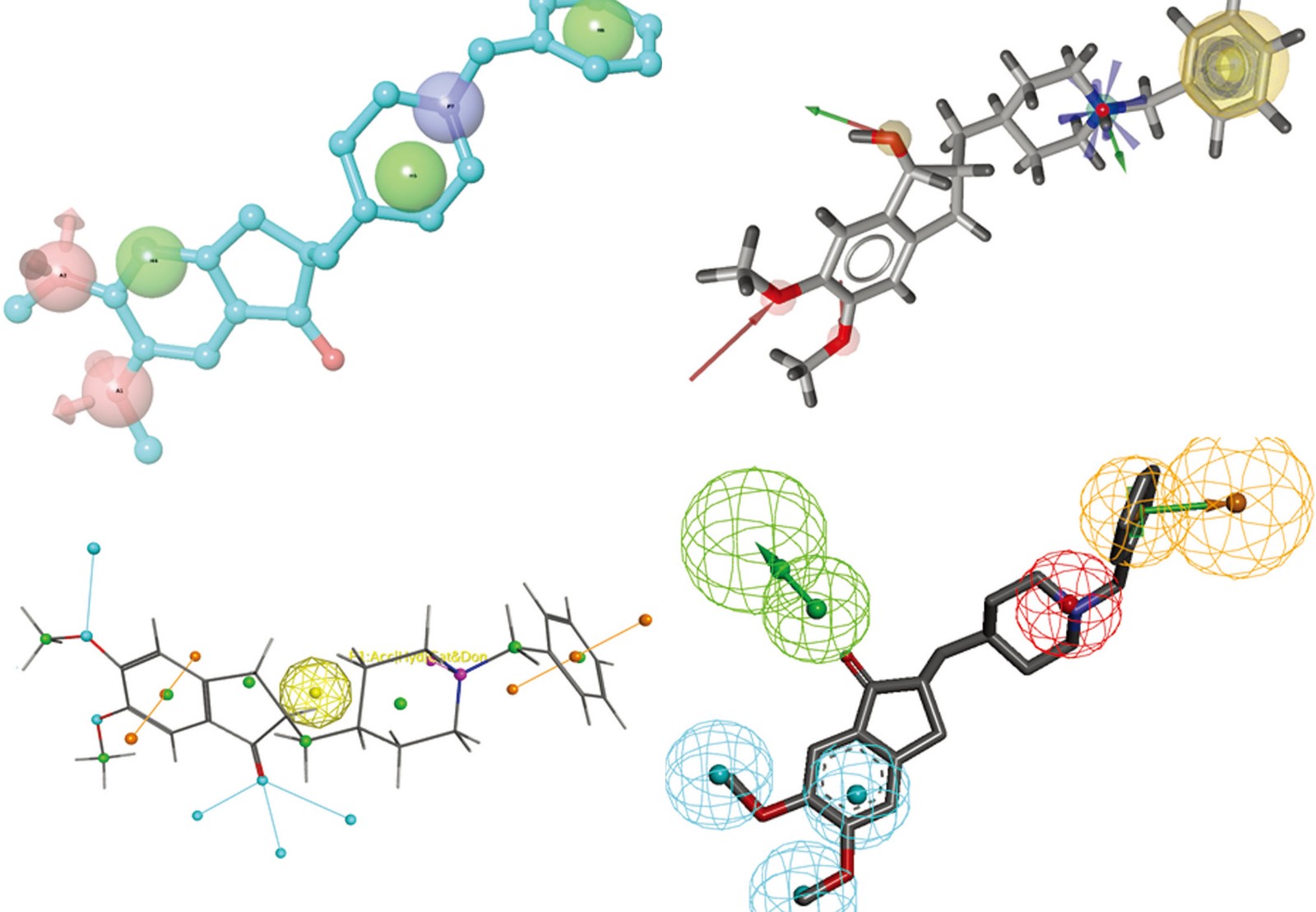
Virtual hit to lead strategies often exploit pharmacophore or molecular docking concepts to identify potential leadcompounds from a virtual-focused library, which is designed around the hit to optimize. For example, a virtuallibrary around the validated fragment can be generated, and then the library can be screened virtually using aconstrained docking strategy to imitate the growing paradigm and maintain the original interaction. A scoringfunction is used to select the best putative optimizations in the hit list. Subsequently, pharmacophore filtersare utilized to extract putative optimizations maintaining the original binding mode.
Creative Biostructure can adopt various ligand-based or receptor-based pharmacophore modelingapproaches based on the available information of the project. In addition, our flexible and customized solutionsallow the use of built pharmacophore models for insilico virtual screening, denovo design, ADME/Tox analysis, lead optimization, multi-target drug design, activity analysis and target identification, etc.
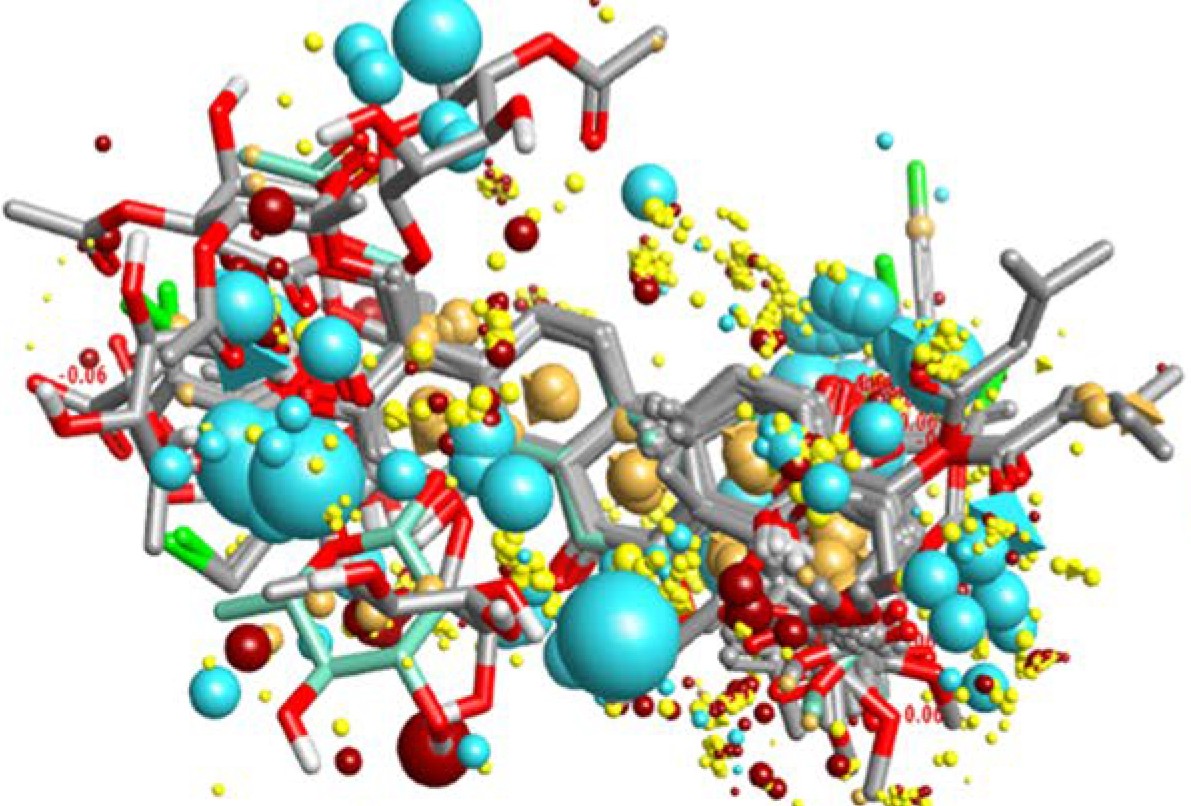
Due to the high cost of obtaining new promising compounds on experimental early drug discovery platforms such as high-throughput screening (HTS), quantitative structure-activity relationship (QSAR) modeling plays a significant role in determining the priority of compounds for synthesis and/or biological evaluation. QSAR model can be utilized for both hit identification and hit to lead, especially the ligand-based drug design process. During hit to lead, a favorable balance between selectivity, potency, and ADME/Tox parameters, which is needed to develop a safe, effective, and novel drug, can be achieved through several optimization cycles. Since no compound needs to be synthesized or tested prior to the computational evaluation, QSAR represents a cost-efficient approach to obtain compounds with desired biological properties.
Creative Biostructure provides QSAR analysis solutions, which can directly reflect the relationship between the biological activity of compounds and their structure, and explore important factors related to the activity of drug molecules, to guide drug discovery projects. The research approaches we can adopt in 3D-QSAR include CoMFA, CoMSIA, Topomer CoMFA, COMBINE, CoMSA, CoRIA, and HQSAR. Continuous integration of new molecular descriptors and machine learning algorithms enables the model to be continuously optimized. The QSAR model we builtcan be used to guide virtual screening and ADME/Tox prediction.
ADME (absorption, distribution, metabolism, and excretion) properties and Tox (toxicity) are essential forunderstanding the efficacy and safety of drug candidates and increasing the probabilities of a successfulprogram. In view of the complexity of current R&D models, in silico ADME-Tox prediction hasbecome the preferred approach for early drug discovery.
Creative Biostructure utilizes advanced computational techniques and machine learning methodsto identify patterns in ADME-Tox data and convert them into knowledge that guides the discovery and optimization oflead compounds. Using the developed predictive models, we can predict a variety of physicochemical properties ofcompounds (such as aqueous solubility, logD, logP, and pKa), drug bioavailability, blood-brain barrier (BBB)permeability, CYP450 metabolite profile, hERG toxicity and more during the early stages of drug development. Inaddition, we can assist in the development of more effective predictive models for specific projects. The predicteddata can be applied to virtual screening and usedas a basis for in vitro ADME-Toxexperiments.
References

Easy access to products and services you need from our library via powerful searching tools