Creative Biostructure provides a variety of experimental structural biology approaches to help elucidate the interaction mechanisms between proteins and other molecules (such as proteins, nucleic acids, small molecule compounds, peptides, etc.). At the same time, we can also use computer-based molecular docking methods to predict the three-dimensional (3D) structures of these interacting partners. We exploit computational docking techniques for various purposes, such as protein-ligand docking, protein-protein docking, protein-nucleic acid docking, antigen-antibody docking, and protein-peptide docking.
Molecular docking is a widely used structure-based drug design technique. We utilize this technology to predict the most probable 3D conformations of small-molecule ligands within target binding sites and to provide quantitative projections of the energy variations involved in the intermolecular recognition event. Moreover, these quantitative estimations of the binding energetics provide rankings for the docked compounds, which is a useful parameter for selecting ligands for experimental profiling. In drug design, the molecular docking method is mainly used to search for small molecules (hits) that have a good affinity with target molecules from small molecule databases.
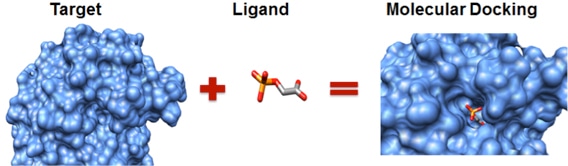
Protein-protein docking is a tool for predicting protein-protein interactions. The precise understanding of protein-protein interactions of disease-related targets contributes to the rational design of therapies. We can use a rigid protein docking algorithm based on the fast Fourier transform correlation technique to realize the protein-protein docking. We can search for all possible binding modes in the translational and rotational space of the protein-protein system, and optimize the binding configurations of the found protein-protein complexes by energy calculation, and then the energy scoring function is utilized to score these binding configurations. This molecular docking strategy has been successfully applied for antigen-antibody docking, which contributes to the affinity maturation of antibodies.
Computational techniques complement experimental approaches in elucidating protein-nucleic acid interactions. Through modeling protein-nucleic acid complex structures at the atomic level, we can yield sufficient information to build a working hypothesis and guide further experimental analyses to identify crucial amino acids or nucleotide residues. We employ computational docking tools to search all possible binding patterns in the translational and rotational space between the protein and nucleic acids (RNA, DNA, or hybrid DNA/RNA), which can undergo large conformational changes upon complex formation.
Creative Biostructure exploits advanced protein-ligand docking software to find the binding conformation of the target and ligand and predict its binding pattern and affinity by calculating the active site, geometry, and energy. Each docking process is a combination of a search algorithm and a scoring function. The drug target structure for docking can be obtained by our experimental structure determination methods or constructed by computational methods. Combined with available compound libraries, we can perform the high-throughput virtual screening of millions of compounds in a short time to provide a hit list for your target protein(s), which is also part of our computer-aided drug design (CADD) platform.

Easy access to products and services you need from our library via powerful searching tools