How to find or design a class of small molecule compounds that bind to the target and change its biological activity under the premise of known target structural information? Structure-based virtual screening (SBVS) is one of the significant means to solve such problems. By using computational methods and the three-dimensional (3D) structural information of the biological target, Creative Biostructure can help you study the underlying molecular interactions involved in protein-ligand binding, and screen out compounds for further optimization in a faster and more economical manner.
SBVS relies on the strategy of docking compounds from libraries against a given target. In addition to the prediction of likely binding conformations, SBVS offers a convenient method of ranking the docked molecules using scoring functions. This classification criterion can be used solely or in combination with other procedures for selecting promising molecules for experimental profiling. SBVS strategies usually rely on the following procedures: (1) Target selection and preparation, (2) compound database selection, (3) molecular docking, and (4) analysis of results. Creative Biostructure's SBVS solution covers all the procedures from the initial stage of the process (including preparation of target structure and compound database) to protein-ligand docking, scoring, as well as postprocessing of top-scoring hits.
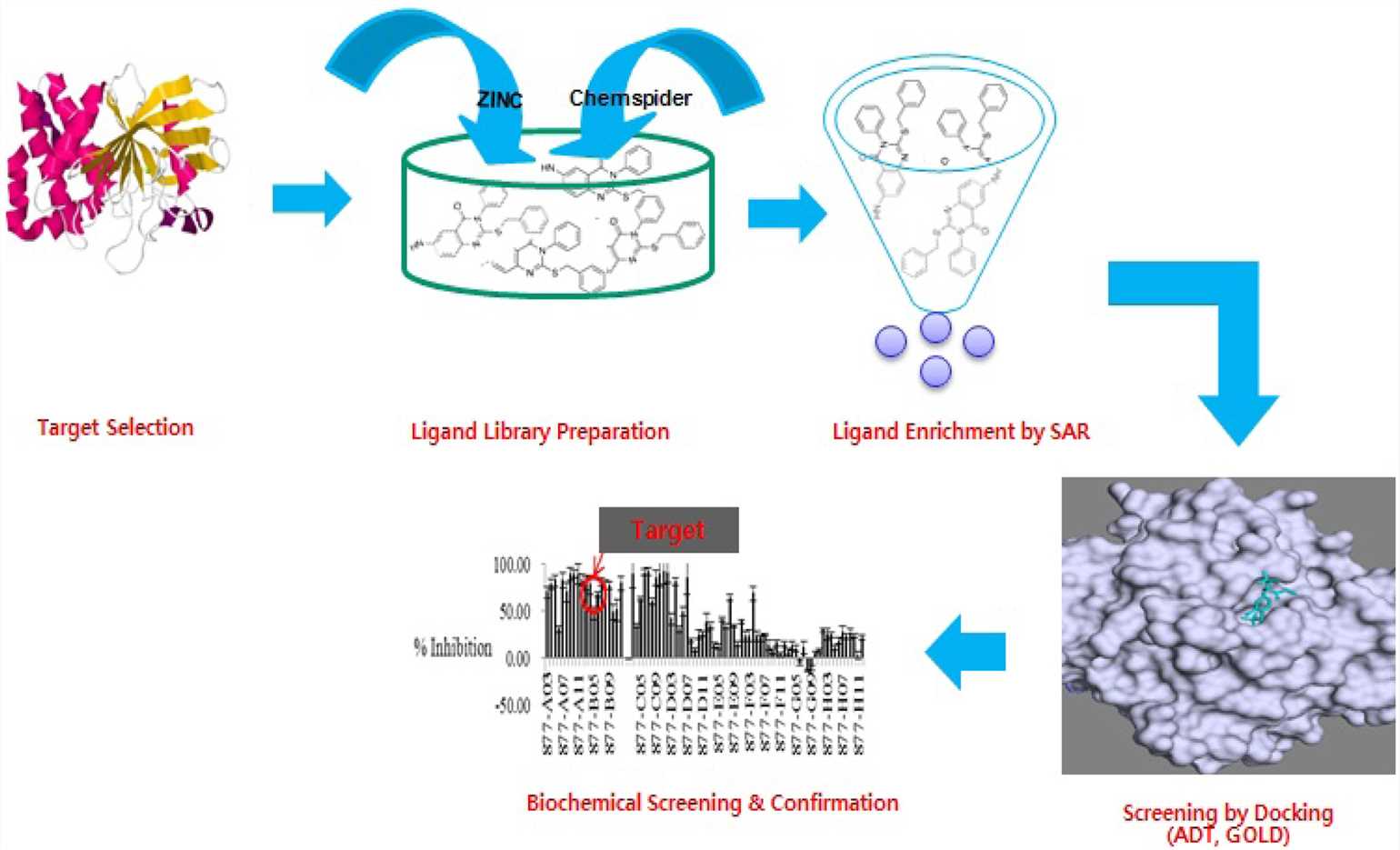 Figure 1. A conceptual figure of structure-based virtual screening. (Reddy R H L.; et al. 2017)
Figure 1. A conceptual figure of structure-based virtual screening. (Reddy R H L.; et al. 2017)
Obtaining the target 3D structure is the starting point of the SBVS project. We can search the PDB database (https://www.rcsb.org/) to find the structure that has been determined or obtain the target structure through homology modeling. In addition, we can utilize mainstream structural biology technologies, including X-ray crystallography, cryo-electron microscopy (cryo-EM), and NMR spectroscopy to provide a one-stop target structure analysis solution from gene to structure. With the aid of computational analysis, we investigate the protein surface from multiple perspectives such as topology, physicochemical properties, and force field to predict the appropriate binding site.
The second important issue is to select and prepare a suitable compound library. Creative Biostructure can perform large-scale SBVS against multiple public and commercial chemical databases and has created in-house databases on the drug-like properties of different chemical species. Moreover, we can also customize the library preparation according to your project requirements.
When the target structure and compound library are prepared, a variety of approaches can be used for virtual screening, including but not limited to, molecular docking and affinity scoring.
At Creative Biostructure, you only need to provide biological information associated with the target, and we can perform SBVS. Through this process, we can screen out a series of hits and corresponding binding modes. Biological assays can be provided to determine the activity of compounds obtained through SBVS. For each SBVS project, we will tailor a cost-effective solution for the specific needs of customers.
Reference

Easy access to products and services you need from our library via powerful searching tools