Molecular dynamics (MD) is a computational simulation of a complex biological system that describes motions, interactions, and dynamics at the atomic level by choosing a "force field" describing all the interatomic interactions and by integrating the Newtonian equations which give position and speed of atoms over time. The significance of computer simulations of macromolecules, which use classical mechanics principles to describe atom behavior, has been widely acknowledged in the field of drug discovery. Creative Biostructure utilizes advanced MD simulation technology to facilitate conformational dynamics studies of protein and protein-ligand, and we can combine MD simulation with molecular docking methods for in silico virtual screening.
The finite conformation models generated by X-ray crystallography, NMR spectroscopy, cryo-electron microscopy, and homology modeling provide valuable insights into the target structure, while MD simulation can offer multiple conformers of the protein structure at a timescale and provide atomic-level information about protein binding thermodynamics and conformational changes under predetermined physiological conditions (such as pressure and temperature). With high-performance computing tools and first-class MD simulation algorithms, we can investigate the dynamics of target proteins, including soluble proteins, membrane proteins, and large molecular machines. In addition, as a CRO in the field of drug discovery, we apply MD simulation to the drug discovery process, including but not limited to:
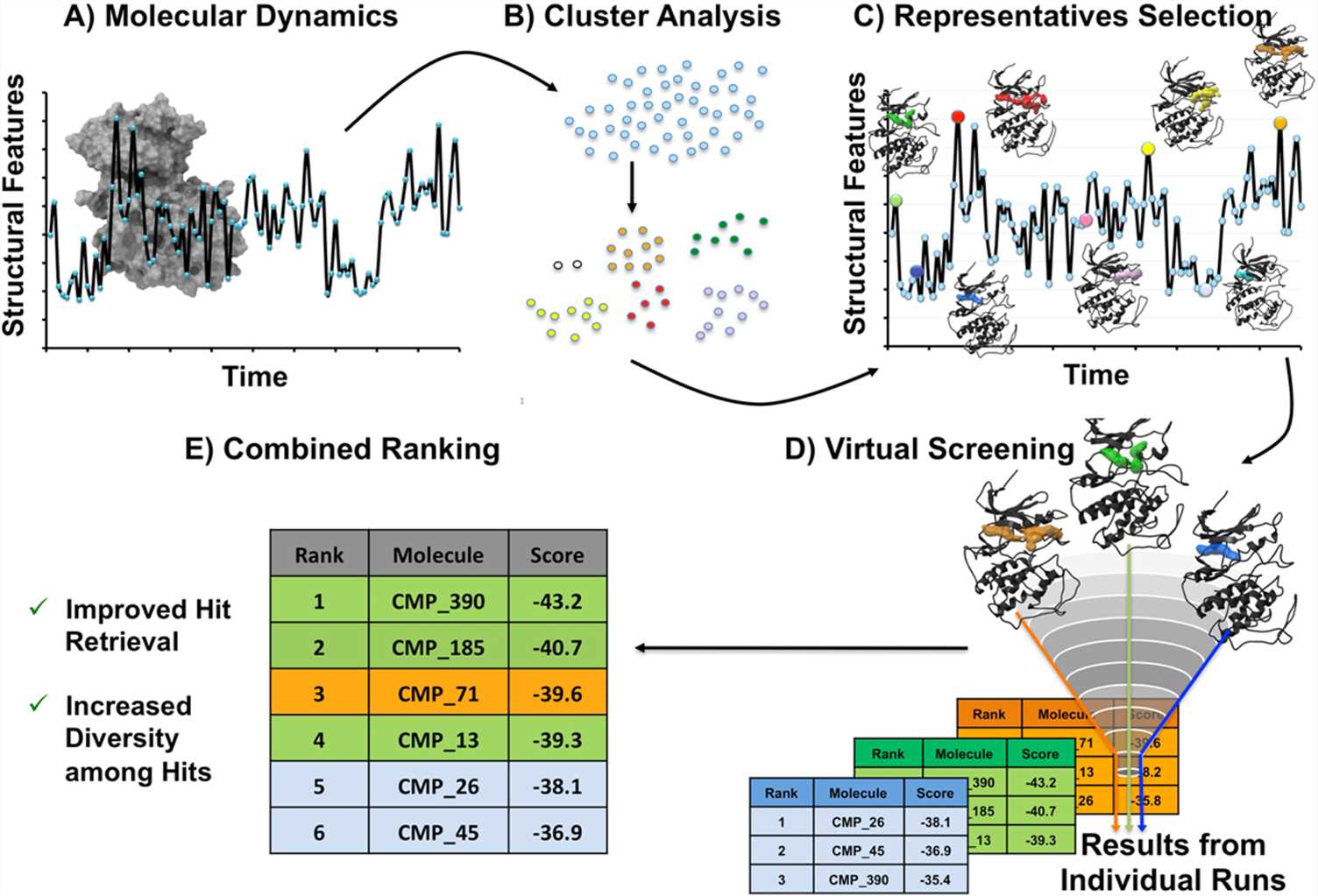 Figure 1. Virtual screening combining MD simulations and molecular docking. (De Vivo M.; et al. 2016)
Figure 1. Virtual screening combining MD simulations and molecular docking. (De Vivo M.; et al. 2016)
Creative Biostructure developed a virtual screening strategy combining molecular docking and MD simulation: 1) Before docking, MD allows a conformational sampling and clustering of a target to account for protein dynamics and the conformational selection by a ligand; 2) MD is also utilized as a second-step filtering process to further validate a protein-ligand complex obtained from docking by determining the stability of the complex from a trajectory, identifying persistent interatomic interactions and by estimating the binding free energy; 3) Finally, by including the induced-fit and solvent effects, we can work with a more realistic model, which may apply even to a membrane-like environment in the case of membrane proteins.
MD simulations and related approaches can help in some key steps of drug discovery, which would offer savings in money and time. MD simulation is incorporated into our computer-aided drug design (CADD) platform to assist in expensive and challenging drug discovery processes.
References

Easy access to products and services you need from our library via powerful searching tools